Molecular Movies Capture Chemistry in Action

This summer, as part of Wellesley’s Summer Research Program, several chemistry students teamed up with Mala Radhakrishnan, associate professor of chemistry, to study the binding processes and properties of microscopic matter. Anik Brinckerhoff ’21 studied the optimal binding properties of the chronic myeloid leukemia drug imatinib as it fastens onto its target protein. Qiao Li DS ’20, focused on the computational design of tighter-binding antimicrobial peptides based on buforin II, a 21 amino acid peptide. Lisa Huang ’21 examined the crowding and conformational dynamics on optimal electrostatic interactions through a case study using the barnase-barstar system.
The students modeled each reaction using computational software, which allowed them to better visualize and understand the complex processes involved. Below is a selection of some of the model videos they produced.
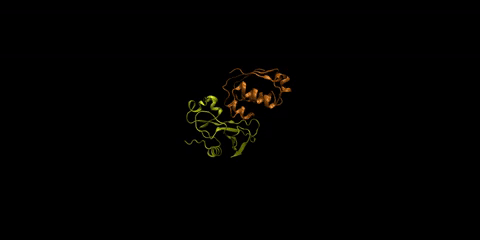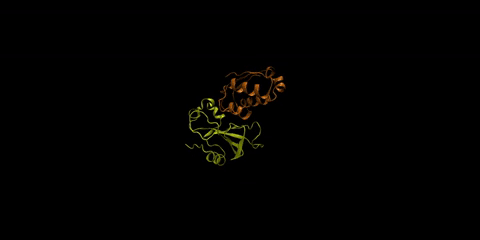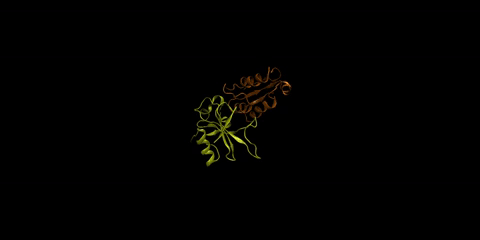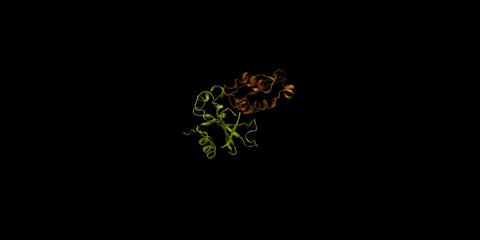
The barnase-barstar complex is one of the most tightly bound natural complexes known. As a bacterial ribonuclease, the function of barnase (in yellow) is to cleave RNA, while barstar (in orange), its intracellular inhibitor, prevents barnase from cleaving the cell’s own RNA before it gets excreted from the cell. Huang is interested in understanding how foreign molecules in the surrounding environment might affect the strong affinity between these two proteins.
“It was fascinating to see how the proteins interact and then be able to determine which residues contribute favorable to binding,” Huang said. “Working in a computational lab solidified the fact that I want to do something similar in the future.”
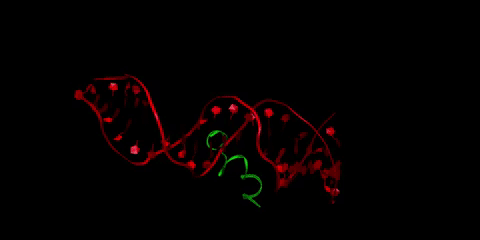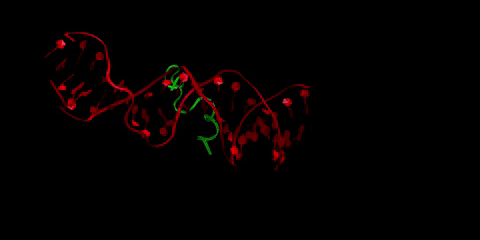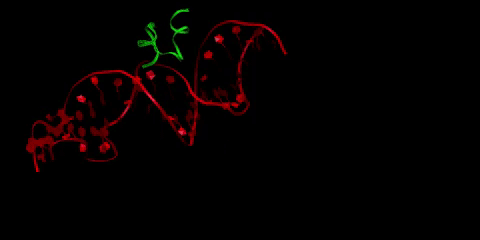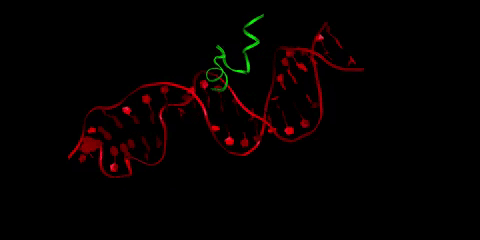
Buforin II (in green) is a type of antimicrobial peptide, a short protein chain that kills bacteria. Here it is pictured bound to DNA (in red), as buforin II is believed to function through binding to bacterial DNA to interfere with the bacteria’s crucial cellular processes. Li is interested in how the crowded cellular environment affects binding in this system, and in designing tighter-binding antimicrobial peptides through collaboration with Radhakrishnan and Donald Elmore, professor of chemistry.
“It’s exciting to use computational modeling because I can obtain more detailed properties of an individual molecule and a higher resolution of molecular interactions than by performing an experimental approach,” Li said. “Working in Professor Radhakrishnan’s lab has opened my eyes to a broader range of research in biochemistry.”

The chronic myeloid leukemia drug imatinib (in green) is shown bound to the ABL kinase domain of its target protein, BCR-ABL kinase (in yellow). Brinckerhoff is studying how mutations in the target protein affect drug-target interactions in this system, and how more optimal electrostatic interactions in this and other rapidly mutating systems might be developed.
“I was fascinated by the intricacy of the challenge of using computational math and Newtonian physics to model the millions of steps needed to approximate the binding complex of a molecule,” Brinckerhoff said. “It was exciting to see it all come to life and see how accurately it reflects the actual real life molecules that impact the treatment of cancer. There’s no better way to learn whether post-graduate research is a good fit than to actually have the opportunity to work in a lab.”
Photo: From left to right, Lisa Huang ’21, Priscilla Oluwakemi Badusi ’21, Anik Brinckerhoff ’21, Mala Radhakrishnan, and Qiao Li DS ’20 review their work in the new science center.



